H1
Setup
Document Rendering
(Detailed document rendering options not shown)
R Packages
library(glue) # formatting strings
library(lubridate) # parsing datetime information
library(kableExtra) # table formatting
library(httr) # or curl, httr, or rvest?
library(xml2)
library(tidyverse) # powerful data processing
theme_set(theme_bw())# white background
setwd(here::here("/home/knut/code/git/_my/work/dis-docs/"))
setwd(here::here("guide/internal"))
Get metadata formats
url_mdfmt="https://doidb.wdc-terra.org/igsnoaip/oai?verb=ListMetadataFormats"
md_fmt <- xml2::read_xml(url_mdfmt, as_html = FALSE, options = "NOBLANKS") %>%
xml_ns_strip() %>%
xml2::xml_find_all(xpath = ".//metadataPrefix", ns = xml_ns(.)) %>%
xml2::xml_text(x = .)
Metadata formats are: oai_dc, igsn.
Get Sets/Metadata Catalogs
url_doi="https://doidb.wdc-terra.org/igsnoaip/oai?"
url_sets="verb=ListSets"
url_catalogs <- sprintf("%s%s", url_doi, url_sets)
catalogs <- xml2::read_xml(url_catalogs) %>%
xml_ns_strip()
setnames <- catalogs %>%
xml2::xml_find_all(xpath = ".//setName", ns = xml_ns(.)) %>%
xml_text()
setspecs <- catalogs %>%
xml2::xml_find_all(xpath = ".//setSpec", ns = xml_ns(.)) %>%
xml_text()
catalogs_df <- tibble(name = setnames, spec = setspecs) %>%
filter(str_detect(.$name, "(?i)Reference quality citations only", negate = TRUE) )
Calculate a data frame of year-intervals
n <- 2009
n_years <- lubridate::year(Sys.Date())
years <- tibble(year = seq(n, n_years))
dates_intervals <- years %>%
mutate(doy_first = sprintf("%s-01-01", year),
doy_last = sprintf("%s-12-31", year))
n_rows <- nrow(dates_intervals)
n_last <- n_rows - 2
dates_intervals %>%
rownames_to_column(var = "#") %>%
slice(c(seq(1,3), seq(n_last, n_rows))) %>%
knitr::kable(row.names = TRUE) %>%
kableExtra::kable_styling(., bootstrap_options = "striped")
| # | year | doy_first | doy_last | |
|---|---|---|---|---|
| 1 | 1 | 2009 | 2009-01-01 | 2009-12-31 |
| 2 | 2 | 2010 | 2010-01-01 | 2010-12-31 |
| 3 | 3 | 2011 | 2011-01-01 | 2011-12-31 |
| 4 | 12 | 2020 | 2020-01-01 | 2020-12-31 |
| 5 | 13 | 2021 | 2021-01-01 | 2021-12-31 |
| 6 | 14 | 2022 | 2022-01-01 | 2022-12-31 |
Goal: Run HTTP GET request for each catalog - year combination.
Step 1/3: Prepare a dataframe of http-calls to verb/method verb=ListSets
url_records = "verb=ListRecords"
url_records_tpl <- glue::glue("{url_doi}{url_records}&metadataPrefix=oai_dc")
http_requests <- catalogs_df %>%
crossing(dates_intervals) %>%
mutate(req = glue("{url_records_tpl}&from={doy_first}&until={doy_last}&set={spec}"))
Step 2/3: Now compose getter function for fetching XML Data via HTTP.
# purrr:partial
status_extract <- compose(httr::status_code, httr::GET)
find_all <- partial(xml2::xml_find_all, xpath = ".//resumptionToken/@completeListSize")
get_count <- compose(possibly(xml2::xml_text, otherwise = "0"),
possibly(find_all, otherwise = "0"),
possibly(xml_ns_strip, otherwise = "0"),
possibly(xml2::read_xml, otherwise = "0"))
Step 3/3: Use the xml getter function to extend the dataframe of URLs with the count of DOIs assigned per year.
This will perform 350 HTTP requests (for all 14 years * 25 catalogs),
outfile_rds <- "../../assets/other/data/igsns_per_year.RDS"
if (file.exists(outfile_rds)) {
#igsns_per_year <- readRDS(outfile_rds)
igsns_per_year <- readRDS(outfile_rds)
} else {
igsns_per_year <- http_requests %>%
mutate(cnt = map(req, get_count))
saveRDS(igsns_per_year, file = outfile_rds)
}
n_rows <- nrow(igsns_per_year)
n_last <- n_rows - 4
igsns_per_year %>%
arrange(year, name) %>%
rownames_to_column(var = "#") %>%
slice(c(seq(1,3), seq(n_last, n_rows))) %>%
knitr::kable(row.names = TRUE) %>%
kableExtra::kable_styling(., bootstrap_options = "striped")
| # | name | spec | year | doy_first | doy_last | req | cnt | |
|---|---|---|---|---|---|---|---|---|
| 1 | 1 | AuScope | ANDS.AUSCOPE | 2009 | 2009-01-01 | 2009-12-31 | https://doidb.wdc-terra.org/igsnoaip/oai?verb=ListRecords&metadataPrefix=oai_dc&from=2009-01-01&until=2009-12-31&set=ANDS.AUSCOPE | |
| 2 | 2 | AuScope Geochemistry Network | LITHODAT.AG | 2009 | 2009-01-01 | 2009-12-31 | https://doidb.wdc-terra.org/igsnoaip/oai?verb=ListRecords&metadataPrefix=oai_dc&from=2009-01-01&until=2009-12-31&set=LITHODAT.AG | |
| 3 | 3 | Australian National Data Service | ANDS | 2009 | 2009-01-01 | 2009-12-31 | https://doidb.wdc-terra.org/igsnoaip/oai?verb=ListRecords&metadataPrefix=oai_dc&from=2009-01-01&until=2009-12-31&set=ANDS | |
| 4 | 346 | MARUM Center for Marine Environmental Sciences | MARUM.HB | 2022 | 2022-01-01 | 2022-12-31 | https://doidb.wdc-terra.org/igsnoaip/oai?verb=ListRecords&metadataPrefix=oai_dc&from=2022-01-01&until=2022-12-31&set=MARUM.HB | 1 |
| 5 | 347 | MARUM Center for Marine Environmental Studies at University of Bremen | MARUM | 2022 | 2022-01-01 | 2022-12-31 | https://doidb.wdc-terra.org/igsnoaip/oai?verb=ListRecords&metadataPrefix=oai_dc&from=2022-01-01&until=2022-12-31&set=MARUM | 1 |
| 6 | 348 | System for Earth Sample Registration | IEDA.SESAR | 2022 | 2022-01-01 | 2022-12-31 | https://doidb.wdc-terra.org/igsnoaip/oai?verb=ListRecords&metadataPrefix=oai_dc&from=2022-01-01&until=2022-12-31&set=IEDA.SESAR | 59771 |
| 7 | 349 | Universität Kiel | UKI | 2022 | 2022-01-01 | 2022-12-31 | https://doidb.wdc-terra.org/igsnoaip/oai?verb=ListRecords&metadataPrefix=oai_dc&from=2022-01-01&until=2022-12-31&set=UKI | |
| 8 | 350 | Universität Kiel - Rechenzentrum | UKI.RZ | 2022 | 2022-01-01 | 2022-12-31 | https://doidb.wdc-terra.org/igsnoaip/oai?verb=ListRecords&metadataPrefix=oai_dc&from=2022-01-01&until=2022-12-31&set=UKI.RZ |
Done with getting data.
Prepare data for plotting
Remove those catalogs that contain a "." in their setSpec attribute.
They are subcatalogs, and often there is only 1 subcatalog for each registrant.
Add a few columns to make calculations and plotting easier.
igsns_per_year_clean <- igsns_per_year %>%
arrange(spec, doy_last) %>%
mutate(cnt = as.integer(cnt)) %>%
filter(str_detect(spec, "\\.", negate = TRUE)) %>% # remove "subcatalogs".
mutate(
year2 = as.Date(glue("{year -1}-12-31")),
spec2 = spec,
spec2 = fct_reorder(spec2, -cnt, sum, na.rm = TRUE),
catalog = as.factor(case_when(
spec2 == "GEOAUS" ~ "GEO AUS",
spec2 == "IEDA" ~ "SESAR",
#spec2 == "GFZ" ~ "GFZ",
TRUE ~ "(other)"
)))
igsns_per_year_max <- igsns_per_year_clean %>%
arrange(spec, doy_last) %>%
filter(!is.na(cnt)) %>%
select(spec, year, cnt) %>%
group_by(spec) %>%
summarize(maxcnt = max(cnt, na.rm = TRUE)) %>%
ungroup() %>%
mutate(max_yearly = as.factor(case_when(
maxcnt >= 10000 ~ ">= 10000",
maxcnt >= 1000 ~ " >= 1000",
TRUE ~ "<= 1000"
)),
max_yearly = fct_relevel(max_yearly, ">= 10000", " >= 1000", "<= 1000")) %>%
select(-maxcnt)
igsns_per_year_clean <- igsns_per_year_clean %>%
inner_join(igsns_per_year_max, by = "spec")
igsns_per_year_sm <- igsns_per_year_clean %>%
filter(catalog == "(other)" )
Plots
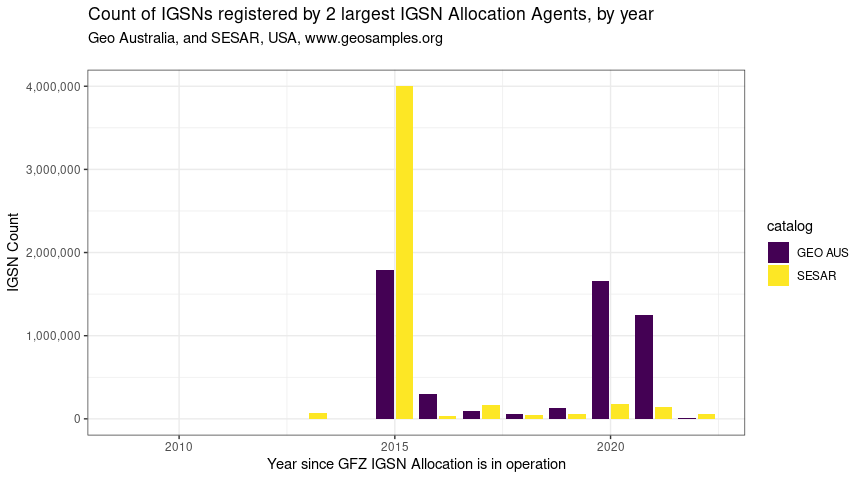
p01_total <- igsns_per_year_clean %>%
filter(catalog != "(other)" ) %>%
filter(year >= 2009) %>%
ggplot(aes(year2, cnt, fill = catalog)) +
geom_bar(stat = "identity", position = "dodge2") +
#theme(legend.position = "none") +
#annotate("text", x = as.Date("2017-12-31"), y = 3000, label = "turquoise = IGSNs of GEOFON records, 2013-2015\nred = IGSNs in all other catalogs") +
scale_x_date() +
scale_y_continuous(labels = scales::label_comma()) +
scale_fill_viridis_d() +
labs(x = "Year since GFZ IGSN Allocation is in operation",
y = "IGSN Count",
title = "Count of IGSNs registered by 2 largest IGSN Allocation Agents, by year",
subtitle = "Geo Australia, and SESAR, USA, www.geosamples.org\n")
p01_total
igsn_svg <- "img/igsn_svg_p01_total.svg"
ggsave(file = igsn_svg, plot = p01_total, width = 10, height = 8)
Smaller IGSN catalogs, Sorted by total number of records
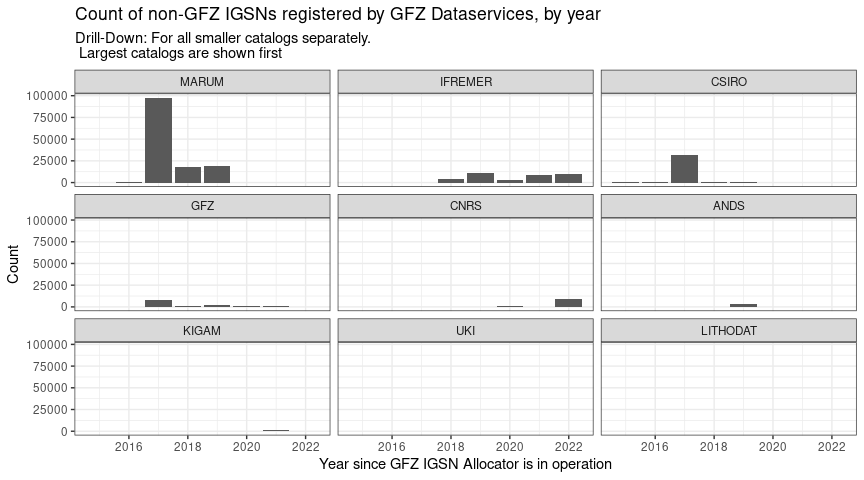
p02_separate <- igsns_per_year_sm %>%
ggplot(aes(year2, cnt)) +
geom_col() +
scale_x_date() +
labs(x = "Year since GFZ IGSN Allocator is in operation",
y = "Count",
title = "Count of non-GFZ IGSNs registered by GFZ Dataservices, by year",
subtitle = "Drill-Down: For all smaller catalogs separately.\n Largest catalogs are shown first") +
facet_wrap(~spec2)
p02_separate
igsn_svg <- "img/igsn_svg_p02_separately_absolute.svg"
ggsave(file = igsn_svg, plot = p02_separate, width = 10, height = 8)
Smaller IGSN Catalogs, with individual Y-Axis scales
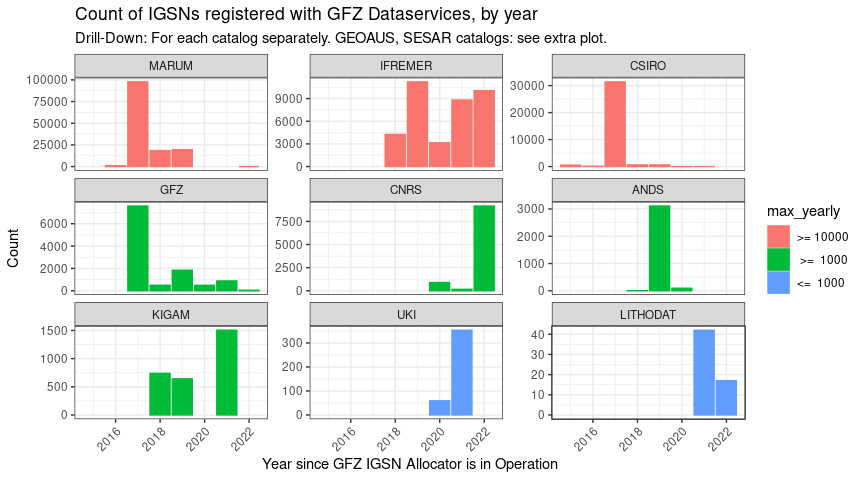
Same plot as before, zoomed in, with individual Y-Axis scales, and subplots sorted by total number of records:
p03_classifc <- igsns_per_year_sm %>%
ggplot(aes(year2, cnt, color = max_yearly, fill = max_yearly)) +
geom_col() +
theme(axis.text.x = element_text(angle=45, hjust = 1)) +
labs(x = "Year since GFZ IGSN Allocator is in Operation",
y = "Count",
title = "Count of IGSNs registered with GFZ Dataservices, by year",
subtitle = "Drill-Down: For each catalog separately. GEOAUS, SESAR catalogs: see extra plot.") +
facet_wrap(~spec2, scales = "free_y")
p03_classifc
igsn_svg <- "img/igsn_svg_p03_with_classific.svg"
ggsave(file = igsn_svg, plot = p03_classifc, width = 10, height = 8)